MICLab researchers develop state-of-the-art software for medical image processing and analysis. We are also devoted to collaborative and reproducible research. See below a list of the latest software and databeses released by our group.
GitHub Tools:
Click the boxes below for more information.
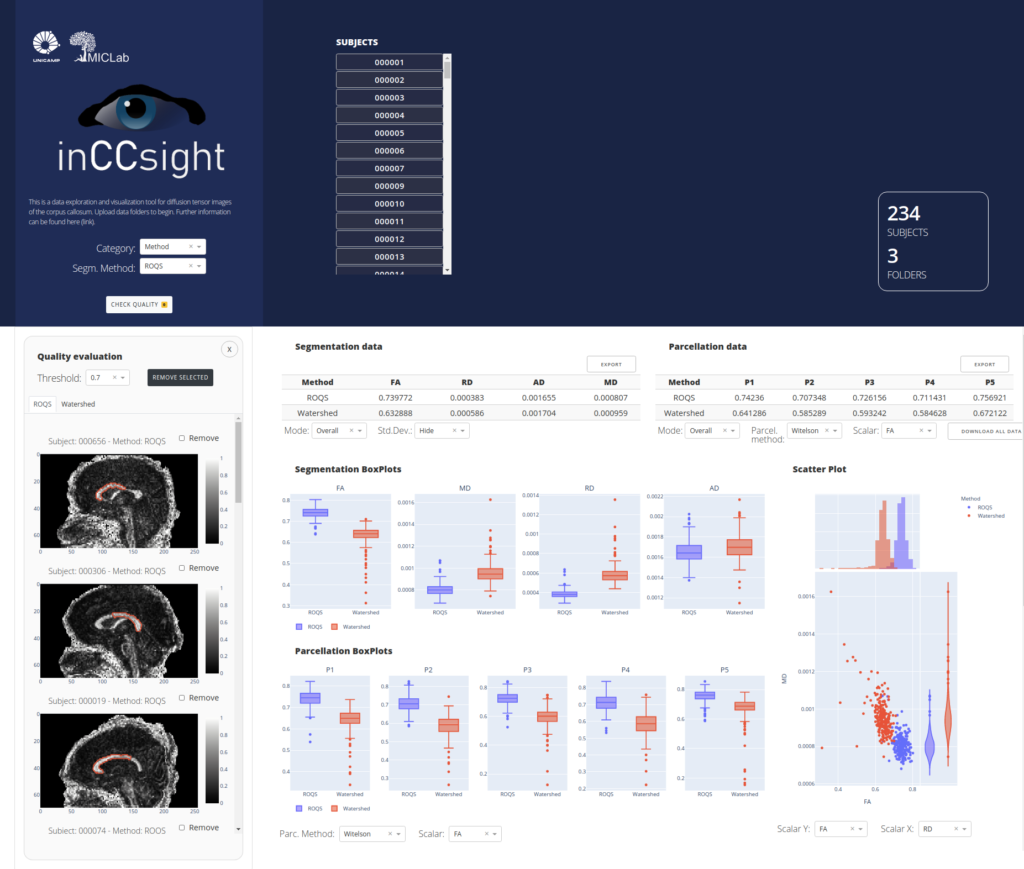
inCCsight is a web-based software for processing, exploring e visualizing data for Corpus Callosum analysis using Diffusion Tensor Images (DTI) implemented in Python/Dash/Plotly.
The software is open source, portable and interactive for analysis of the corpus callosum in DTI individually or in groups, implementing different techniques available for segmentation and installment and proposing relevant metrics for comparing and evaluating the quality of these procedures in a web application.
Extended 2D Consensus Hippocampus Segmentation (e2dhipseg) – beta
Introduction
This work implements the segmentation method proposed in the paper Hippocampus segmentation on epilepsy and Alzheimer’s disease studies with multiple convolutional neural networks published in Heliyon, Volume 7, Issue 2, 2021 (https://www.sciencedirect.com/science/article/pii/S2405844021003315)
A masters dissertation related to this work has been published: Deep Learning for Hippocampus Segmentation (http://repositorio.unicamp.br/handle/REPOSIP/345970)
This repository is also the official implementation for the short paper Extended 2D Consensus Hippocampus Segmentation presented at the International Conference on Medical Imaging with Deep Learning (MIDL), 2019
Authors: Diedre Carmo, Bruna Silva, Clarissa Yasuda, Leticia Rittner, Roberto Lotufo
This code is mainly intended to be used for others to be able to run this in a given MRI dataset or file. Notice we published a standalone release with a minimal GUI for ease of prediction. Any problem please create an issue and i will try to help you.
Thank you!
Minimum Requirements
At least 8GB of RAM Ubuntu 16.04 or 18.04, might work in other distributions but not tested
Having a GPU is not necessary, but will speed prediction time per volume dramatically (from around 5 minutes in CPU to 15 seconds in a 1060 GPU).
Software Requirements
If you are using the binary release, no enviroment setup should be necessary besides FLIRT, if used.
To run this code, you need to have the following libraries installed:
python3
pytorch >= 0.4.0 and torchvision
matplotlib
opencv
nibabel
numpy
tqdm
scikit-image
scipy
pillow
h5py
FLIRT – optional if you want to use the automated volume registration, please do use it if your volumes are not in the MNI152 orientation, or you will get blank/wrong outputs\
You can install pytorch following the guide in in https://pytorch.org/. If you plan to use a GPU, you should have the correct CUDA and CuDNN for your pytorch installation. For CPU only installation on Windows use the following commands:
- pip install torch==1.3.0+cpu -f https://download.pytorch.org/whl/torch_stable.html
- pip install torchvision==0.2.2.post3
A Windows standalone executable can be compiled with:
- pyinstaller e2dhipse_windows.spec
FLIRT can be installed following https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FslInstallation/Linux (for Windows executables are aleady included) The other requirements can be installed with pip3
COEdet is a Linux graphical interface tool allowing for segmentation of the lung and COVID-19 related findings in CT images.
Modified EfficientDet Segmentation (MEDSeg)
Official repository for reproducing COVID segmentation prediction using our MEDSeg model.
The publication for this method, Multitasking segmentation of lung and COVID-19 findings in CT scans using modified EfficientDet, UNet and MobileNetV3 models, has been published at the 17th International Symposium on Medical Information Processing and Analysis (SIPAIM 2021), and won the “SIPAIM Society Award”. http://dx.doi.org/10.1117/12.2606118
The presentation can be watched in YouTube: https://www.youtube.com/watch?v=PlhNUD0Y4hg
siamxt is a python morphological toolbox focused on a data structure known as max-tree, which allows image processing based on size and shape of structures. Our group currently uses it to develop brain and carotid segmentation on magnetic resonance images.
This is an alpha version of the simple max-tree toolbox (siamxt). The major difference to the iamxt toolbox is that it doesn’t depend on OpenCV, SWIG, OpenMP and it doesn’t come with the drawing methods. The toolbox was implemented in Python and the critical functions were implemented in C++ and wrapped in Python using SWIG. I run the SWIG files on my machine and I provide its output directly to you, so you don’t have to install SWIG on your machine. We have a different max-tree structure that is more suitable for array processing, and allows a fast development of new methods with a reasonable processing time. Our main goal in providing this toolbox is to spread the max-tree data structure further among the scientific community, investigate it further, and develop new tools that may be applied to solve real signal, image processing, and pattern recognition problems. This toolbox works for both 2D and 3D images of type uint8 and uint16.
There is a public SoftwareX article describing the toolbox. The article is available for download here: http://www.sciencedirect.com/science/article/pii/S2352711017300079 If you use this toolbox on your work, please cite the this article.
– Souza, Roberto, et al. “iamxt: Max-tree toolbox for image processing and analysis.” SoftwareX 6 (2017): 81-84.
Additional Resources:
Click the boxes below for more information.
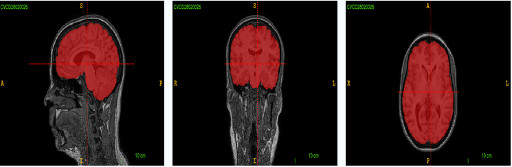
Calgary-Campinas 359 is a public dataset containing T1 weighted brain magentic resonance images coming from three different scanner vendors (Philips, GE, Siemens) and two magnetic field intensities (1.5T and 3.0T).
The dataset has grown over the years and it was moved to: link
General Information
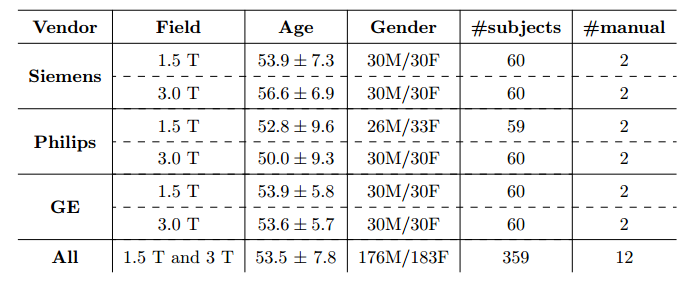
The public dataset we have developed consists of T1 volumes acquired in 359 subjects on scanners from three different vendors (GE, Philips, and Siemens) and at two magnetic field strengths (1.5 T and 3 T). Data was obtained using T1-weighted 3D imaging sequences (3D MP-RAGE (Philips, Siemens), and a comparable T1-weighted spoiled gradient echo sequence (GE)) designed to produce high-quality anatomical data with 1mm^3 voxels. Older adult subjects were scanned between 2009 and 2016.
Smaller, private datasets in Campinas, São Paulo, Brazil and Calgary, Alberta, Canada were used to randomly select the 359 subjects, except for the Philips 1.5 T data where only 59 subjects were available. Age and gender for all subjects was known, however information about subject ethnicity was not available. The Philips 3 T data (60 subjects) were collected in Campinas and the remaining data came from Calgary data sources (299 subjects). The Calgary-Campinas-359 (CC-359) dataset, including the original Nifti files, the consensus masks generated for all subjects using both the STAPLE algorithm and a supervised classification procedure and the manual segmentations of twelve subjects. Detailed information about the acquisition parameters, such as echo time, repetition time, etc., can be provided upon request.
When using this dataset, please cite the following article.
Download information
The dataset file name conventions are the following:
- Original: ____.nii.gz
- STAPLE probability mask: _____staple.nii.gz
- “Silver standard” binary mask: _____ss.nii.gz
- Manual segmentation: _____manual.nii.gz
People interested in downloading CC-359 dataset have to follow the link and fill the formulary: CC-359. Any questions about the dataset, please contact: roberto.medeirosdeso@ucalgary.ca
Throughout the years MICLab has gained Brazlian national news attention and has published in many prestigious journal. See some MICLab videos explaining our research, some are in Portuguese with English subtitles.


