
Segmentation
Segmentation of brain structures has always been of great interest to the medical community. It provides important information on the shape and volume of structures, which can be used to characterize populations and evaluate the progression of diseases. This process is crucial for both clinical and research applications. In both cases, a large number of images must be processed in a robust and reliable manner. Since manual segmentation is a time-consuming task, several automated methods have been proposed in recent years.
The use of deep learning in medical imaging has been growing over the past few years, addressing these issues and providing fast and reliable segmentation methods. In our lab, we leverage deep learning to develop state-of-the-art, reliable, and fast segmentation models. Our methods are designed to run on T1, T2, diffusion MRI, and various other MRI sequences. Some of the successful brain structure segmentation cases our team has developed include the Thalamus [1], Hypothalamus [2], and Corpus Callosum [3], Hippocampal [4], and Skull Striping [5]. Our state-of-the-art results in these areas are made available through public tools.
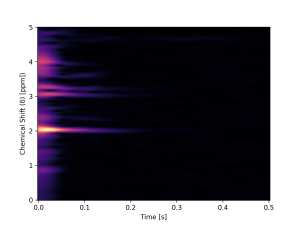
Magnetic Resonance Spectroscopy (MRS) is a technique used to obtain the concentration of metabolites in the brain. This is done by acquiring a spectrum whose peaks represent each component. As the signal has a low signal-to-noise ratio (SNR), it is necessary to collect many spectra (transients), making it a time-consuming technique, which complicates its clinical application. Therefore, it is crucial to balance data quality with minimizing scan duration.
On the other hand, reconstruction is a commonly used term in the literature, describing the process of recovering information lost through acceleration techniques.
Within this context, MICLab developed an innovative approach to reconstruct GABA spectra. This approach, a Deep Learning model named Spectro-ViT, utilizes a vision transformer combined with the spectrogram representation of MRS signals. Spectro-ViT reconstructs data using only one-quarter of the conventional full-data amount, achieving quality metrics comparable to the full-data acquisition pipeline and the best concentration results in the literature.
Our Spectro-Vit pipeline is on GitHub.

Spectroscopy
MRI
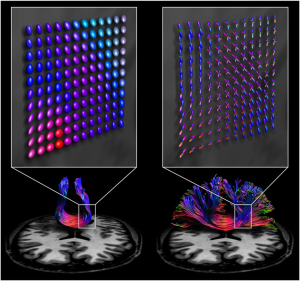
Diffusion MRI (dMRI) is crucial for various tasks, such as surgical planning and monitoring disease progression. Generally, there are two main types of acquisition: single-shell low angular resolution diffusion imaging (LARDI) and high angular resolution diffusion imaging (HARDI). LARDI acquisition is fast and widely used in clinical contexts. However, it faces significant challenges in reconstructing complex fibers, such as those that cross, kiss, fan, or bend. On the other hand, HARDI acquisition can reconstruct these complex fibers with greater accuracy, facilitating specialists’ interpretation. However, the acquisition time is longer, making this technique clinically impractical in many cases.
Given the importance of obtaining high-quality data with a clinically viable acquisition time, MICLab develops advanced dMRI reconstruction methods. These methods are designed to handle fiber complexity, providing an effective solution for reconstructing complex fibers in clinical contexts [1, 2].
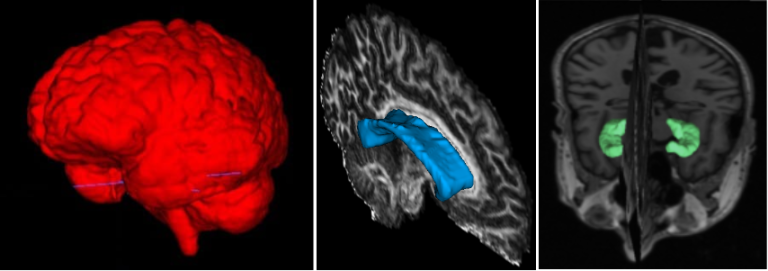
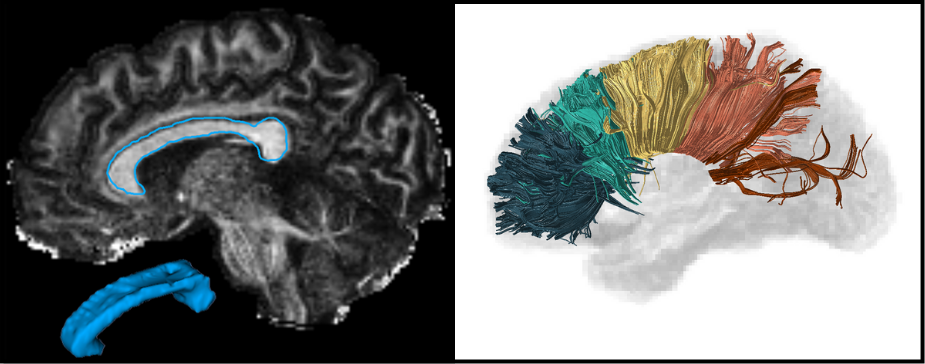
The corpus callosum (CC) is the largest white matter structure in the brain, composed of axonal fibers that play an important role in interhemispheric cerebral communication. Therefore, its characterization is crucial for studying several neurological diseases and neuropsychiatric disorders.
The CC segmentation is necessary when analyzing changes in this structure [1]. We developed various methods to perform this task directly in Diffusion Tensor Imaging (DTI), including a volumetric segmentation of the whole structure using Deep Learning techniques [2]. The subdivision of the CC into a given number of regions, known as parcellation, it is also important and facilitates the analysis of their properties and brain connectivity. Our group proposed several automated data-driven CC parcellation methods using DTI [3,4]. We also aim to extract the CC signature from MRI, which could be useful for population comparison, monitoring changes, or identifying abnormalities.
Most of our proposed methods are available through an open-source software called inCCsight [5]. With its user-friendly interface and advanced features, inCCsight is a powerful choice for DTI data analysis related to the CC.
Magnetic Resonance Images
Imaging
Spectral imaging unlocks a new dimension in Computer Vision, allowing the analysis of scenes across the electromagnetic spectrum. Unlike traditional RGB imaging, which typically provides information in three spectral bands (red, green, blue), hyperspectral imaging divides the electromagnetic spectrum into many more bands, and ranges beyond the visible spectrum.
Because different materials interact in different manners depending on the incident radiation, their unique response across the spectrum can be used to characterize and identify them. By integrating spatial and spectral data, spectral imaging addresses two critical questions in analysis: “What” is present and “Where” it is located. This dual capability enables simultaneous differentiation of tissues or materials and delineation of regions sharing similar spectral characteristics.
This technology was first explored in Remote Sensing, but, since then, has been appropriated by many other fields, including Medicine. In medical applications, it assists surgeons in identifying tumors and evaluating tissue oxygen levels, thereby improving diagnostic precision and surgical outcomes.
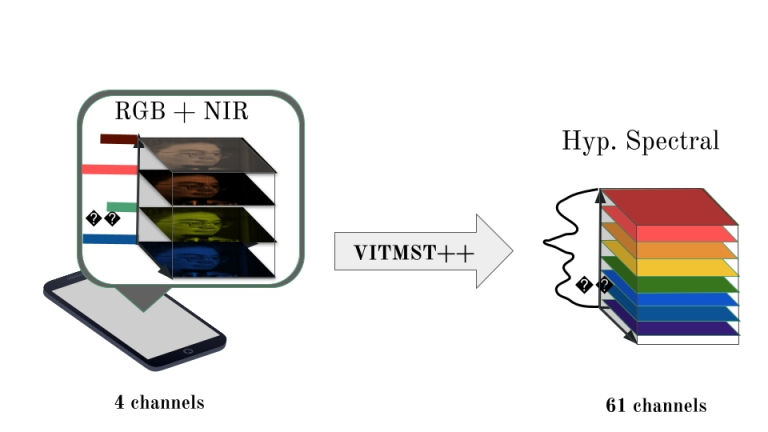
Our research focuses on advancing spectral image acquisition and processing. We develop cost-effective multispectral systems, refine hyperspectral image segmentation techniques, and explore smartphone-based hyperspectral image reconstruction to broaden its applications in healthcare and beyond.
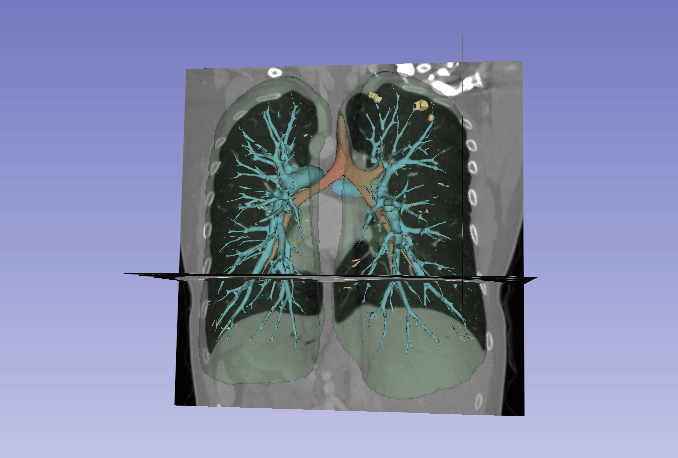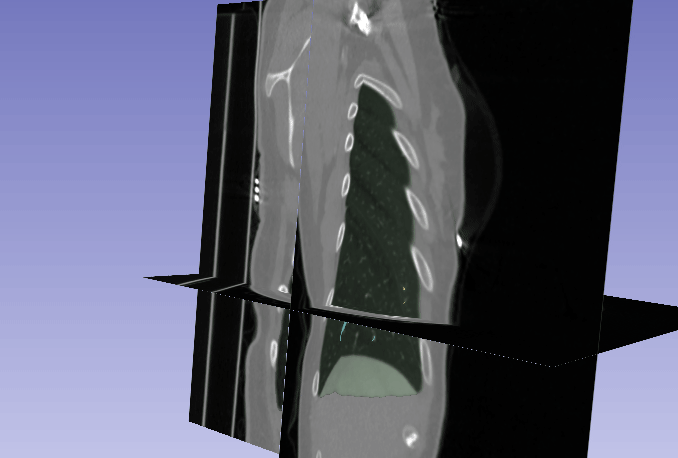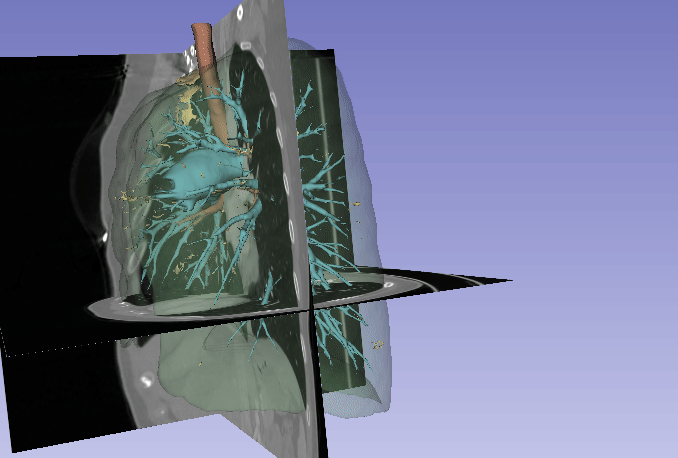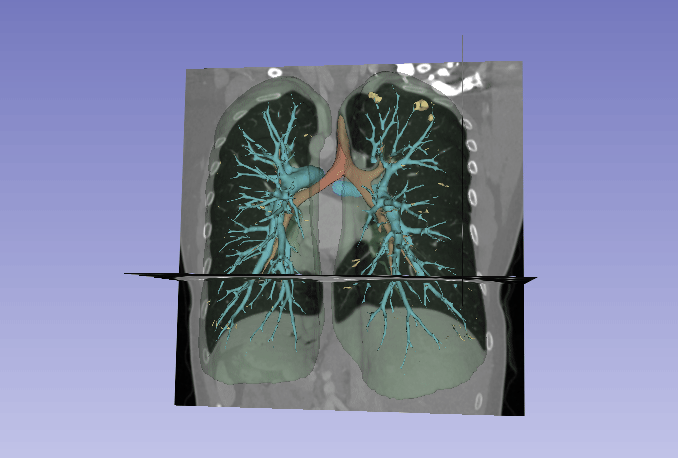
Automated segmentation of the lung and its structures, such as lung lobes, findings, vessels, and airways, in medical images is a crucial task with various important applications in radiology and medicine. Segmentation methods can aid in diagnosing, monitoring, and treatment planning of lung diseases, including understanding emerging diseases such as COVID-19.
Despite technological advances, lung segmentation in computed tomography (CT) images faces challenges such as anatomical variations among patients, noise and artifacts in medical images, and morphological changes caused by lung diseases. Conditions like lung cancer, with nodules, or COVID-19, with consolidations and opacities, add to the complexity of accurate lung segmentation.
In response to these challenges, our research team has developed MEDPSeg, an open-source lung structure and findings segmentation tool designed specifically for robust automated segmentation of lungs containing disease-related findings. MEDPSeg is trained on thousands of CT scans and provides comprehensive segmentation of the lung, lung lobes, opacities, consolidations, airways, and pulmonary arteries. MEDPSeg is lightweight and requires only 6 GB of VRAM to run.
Explore MEDPSeg and its capabilities at GitHub – MICLab-Unicamp/medpseg.